# A tibble: 32 × 1
preference
<chr>
1 cat
2 dog
3 dog
4 dog
5 dog
6 cat
7 dog
8 dog
9 cat
10 dog
# ℹ 22 more rowsTest of and associations between categorical variables
Lecture 6
Iain R. Moodie
BIOB11 - Experimental design and analysis for biologists
Department of Biology, Lund University
2025-04-02
Tests of proportion
Tests of proportion
Could a proportion have been observed under a null hypothesis?

Tests of proportion
Could a proportion have been observed under a null hypothesis?
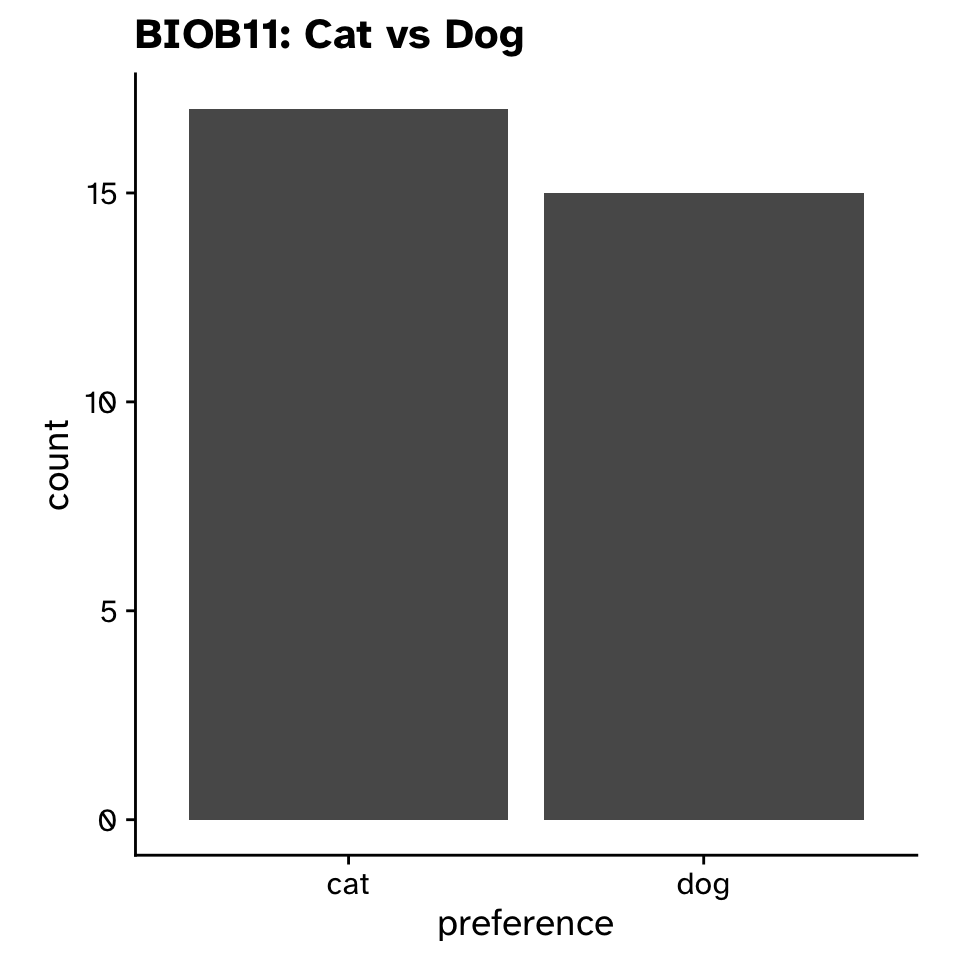
Tests of proportion
Could a proportion have been observed under a null hypothesis?
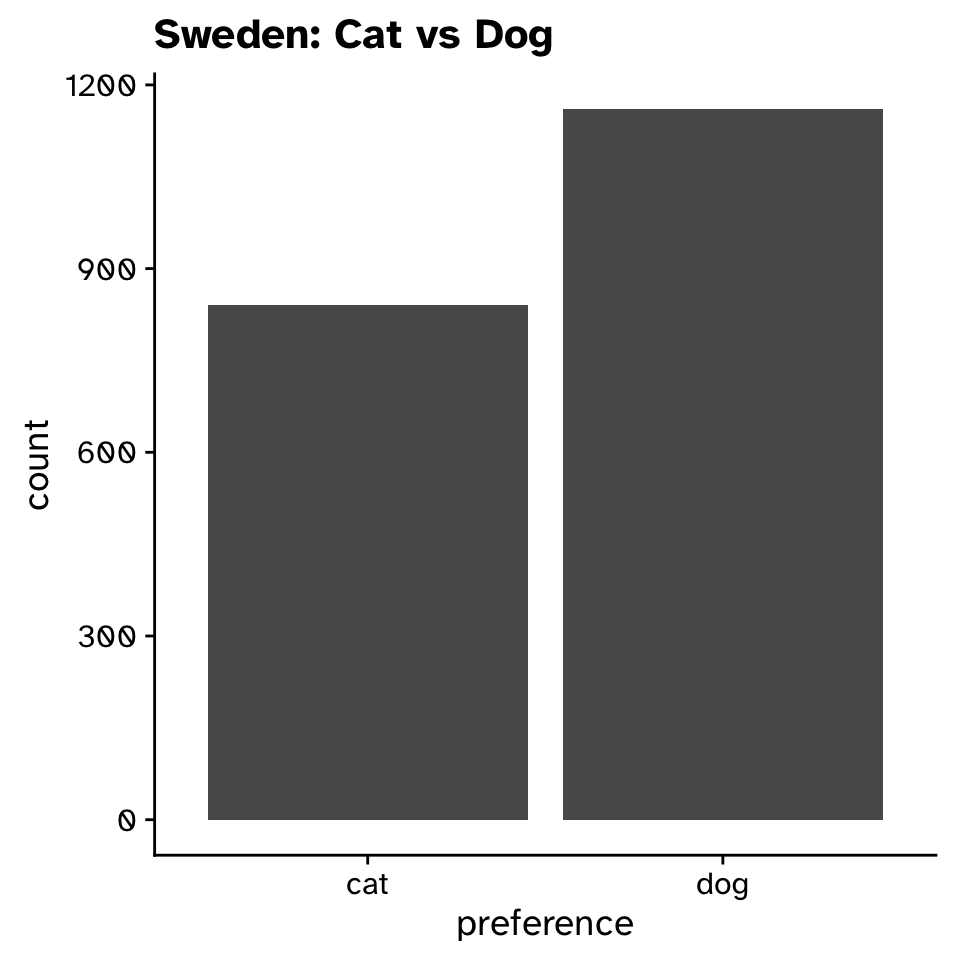
Tests of proportion
Could a proportion have been observed under a null hypothesis?
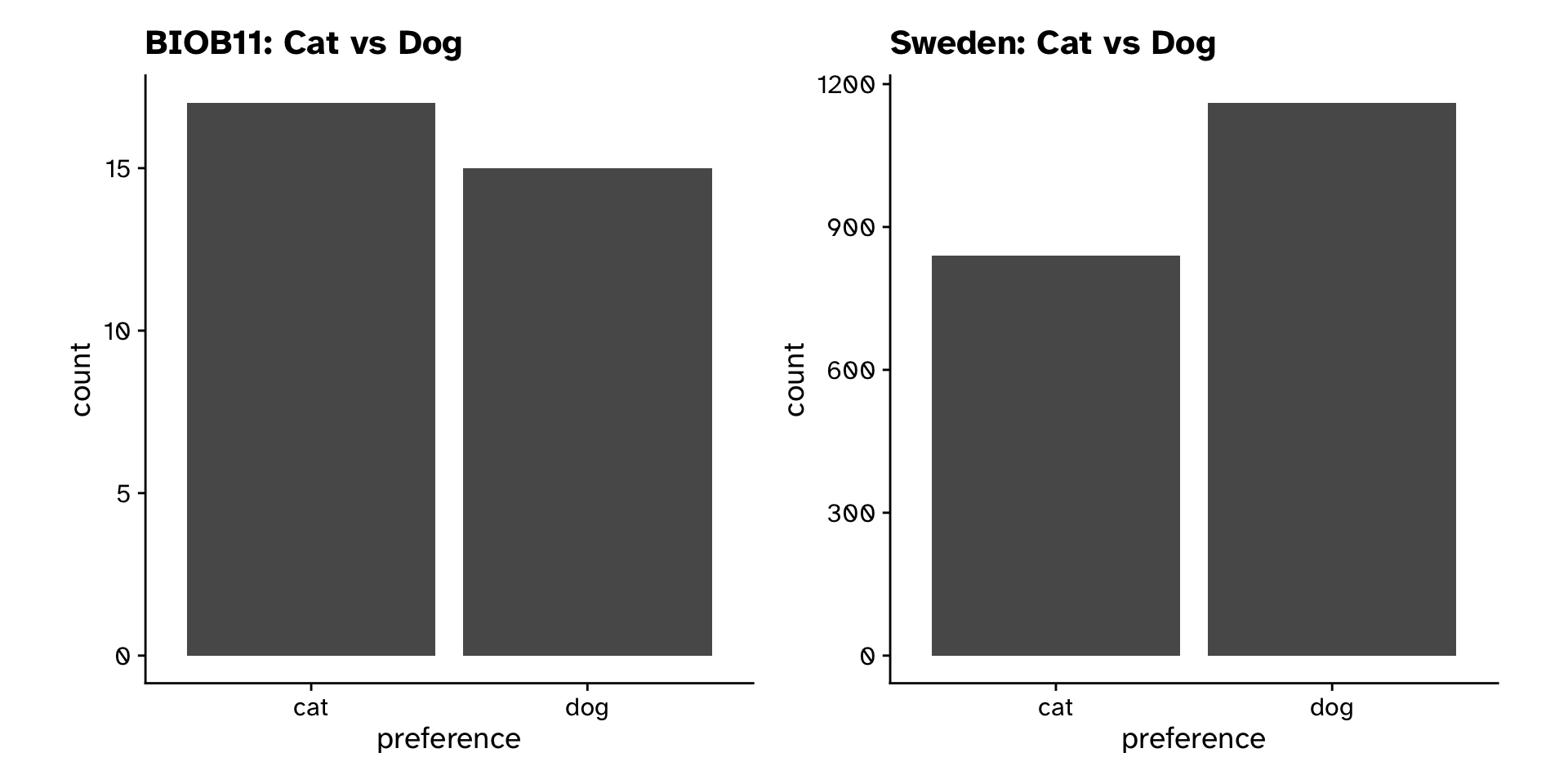
Tests of proportion
Could a proportion have been observed under a null hypothesis?
- Null hypothesis:
- BIOB11 does not differ in their preference for cats vs dogs from the Swedish population
- Alternative hypothesis:
- BIOB11 does differ in their preference for cats vs dogs from the Swedish population
Tests of proportion
Could a proportion have been observed under a null hypothesis?
Tests of proportion
Could a proportion have been observed under a null hypothesis?
Calculate the test statistic:
Tests of proportion
Could a proportion have been observed under a null hypothesis?
Generate a null distibution:
Tests of proportion
Could a proportion have been observed under a null hypothesis?
Visualise the null:
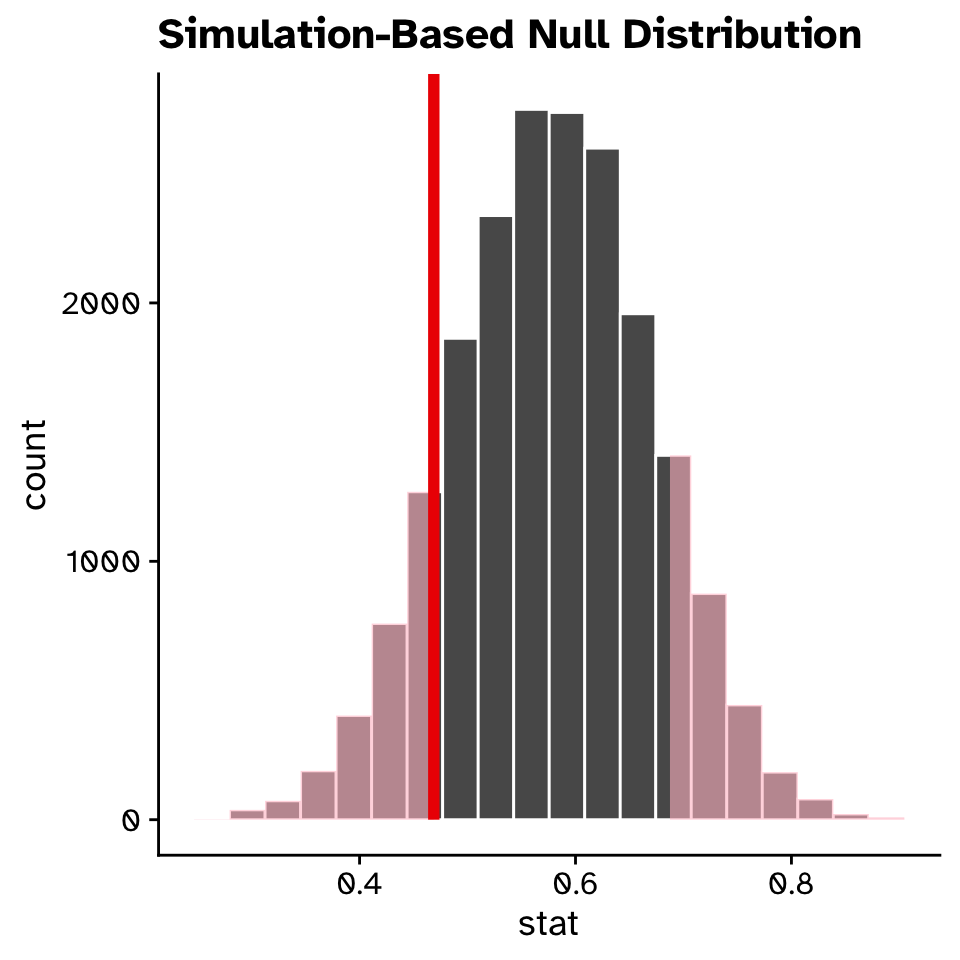
Tests of proportion
Could a proportion have been observed under a null hypothesis?
Calculate the p-value:
\(\chi^2\) Goodness of fit
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
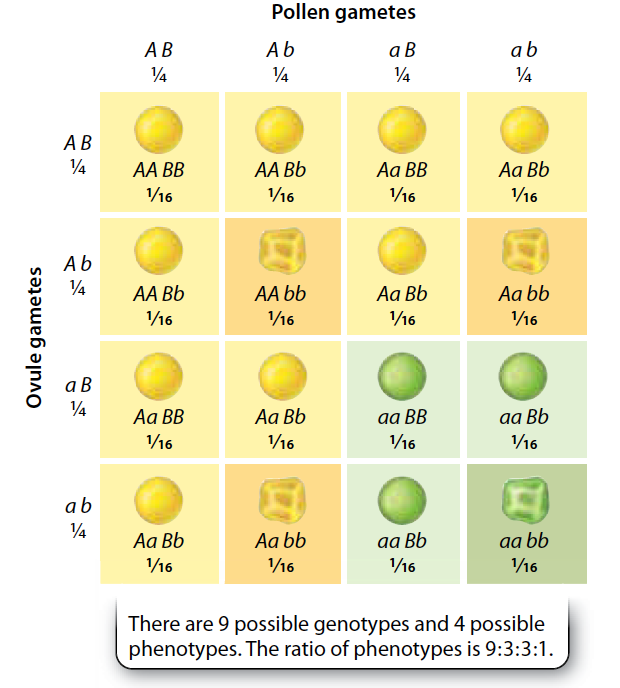
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
- Null hypothesis:
- The sample came from the hypothesised distribution
- The sample distribution is not different from the hypothesised distribution
- Alternative hypotheis:
- The sample came from a different distribution to the one hypothesised
- The sample distribution is different from the hypothesised distribution
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
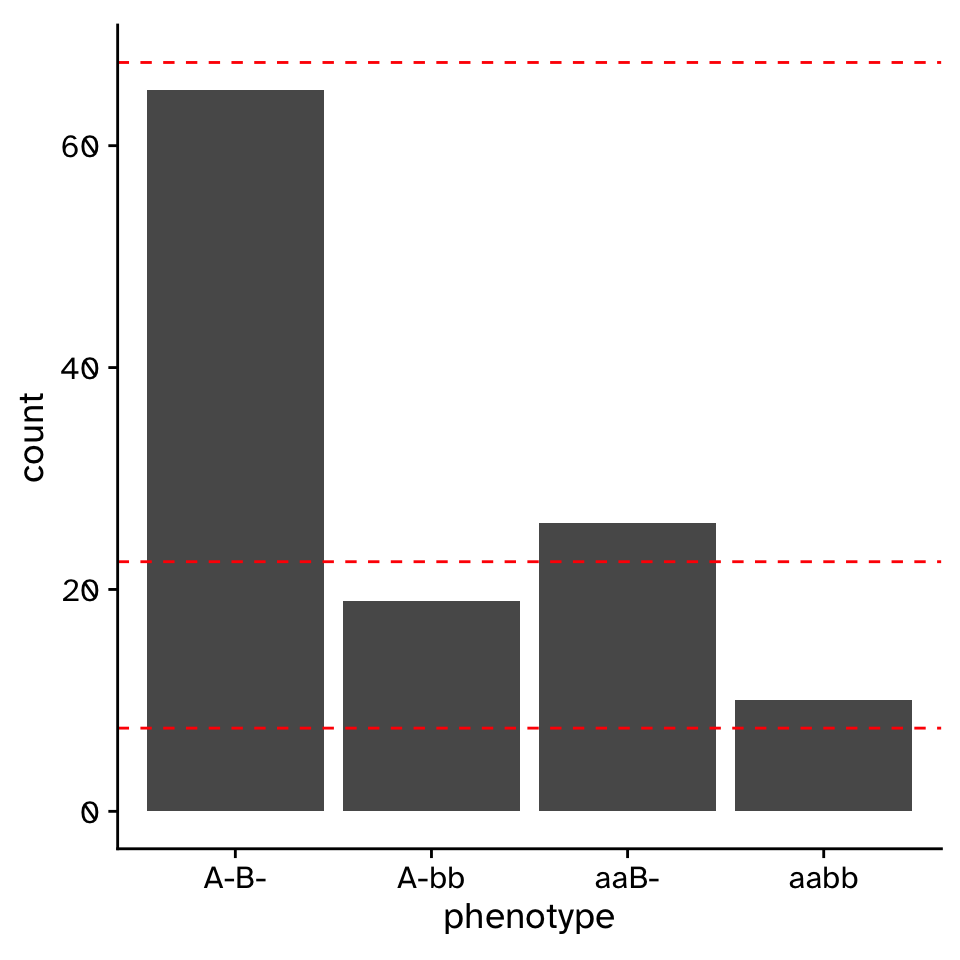
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\[ \chi^2 = \sum \frac{(Observed_i - Expected_i)^2}{Expected_i} \]
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
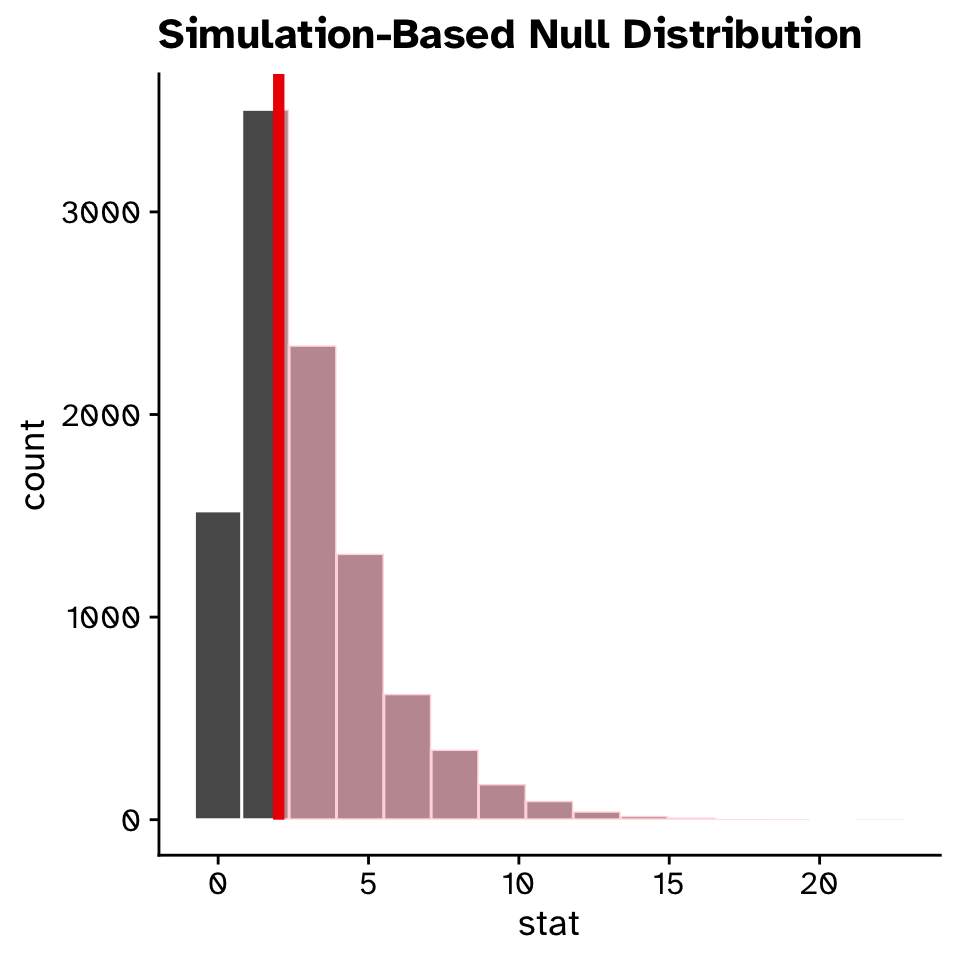
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
\(\chi^2\) Goodness of fit
Does the observed data differ from an expected distribution?
- Fail to reject null hypothesis.
- The distribution of phenotypes is compatible with mendelian inheritance.
\(\chi^2\) Test of independence
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
- Null hypothesis:
- The two categorical variables are not associated with each other
- The two categorical variables are independent
- Alternative hypothesis:
- The two categorical variables are associated with each other
- The two categorical variables are not independent
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
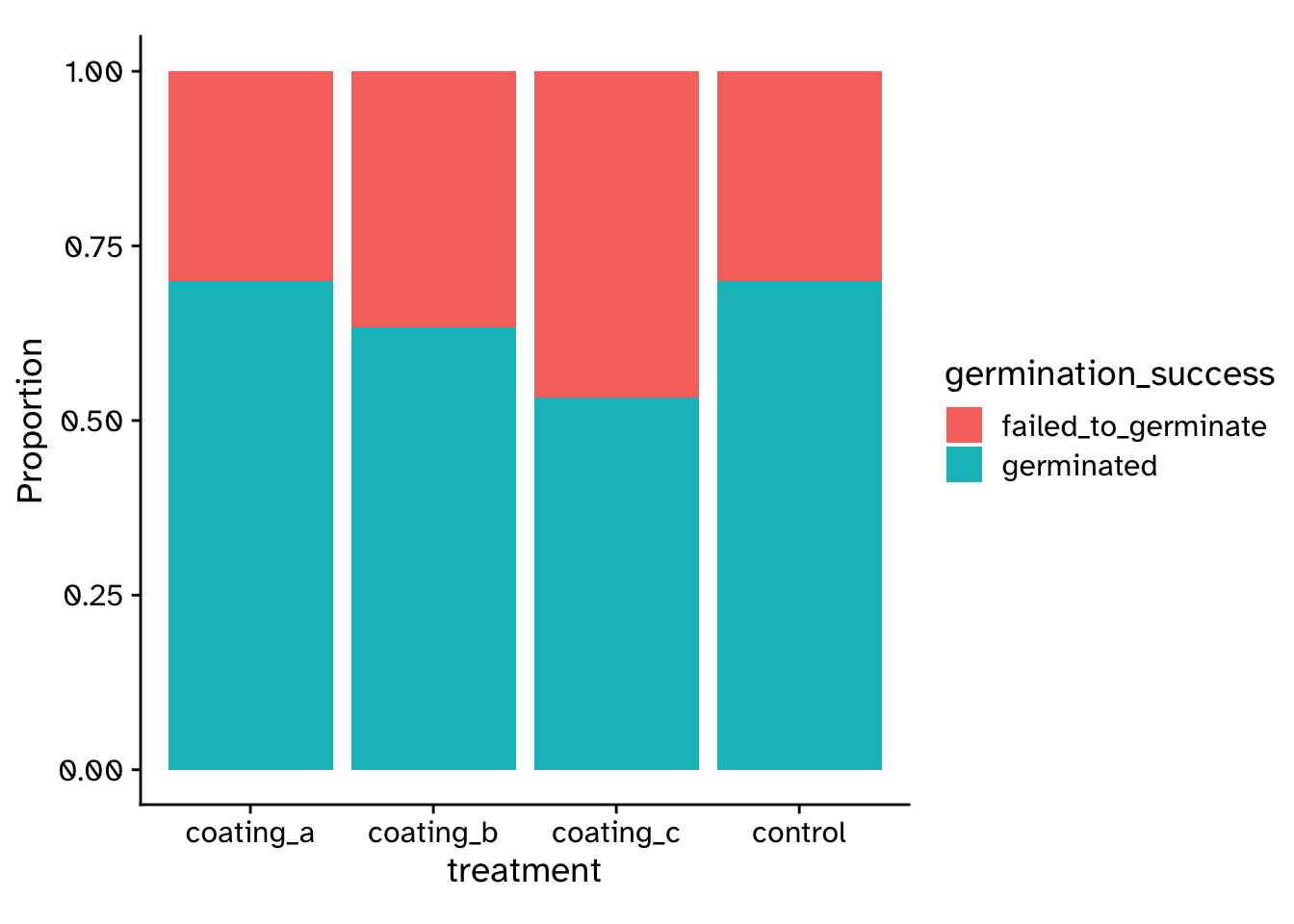
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
# A tibble: 120 × 2
treatment germination_success
<chr> <chr>
1 coating_a germinated
2 coating_a germinated
3 coating_a germinated
4 coating_a failed_to_germinate
5 coating_a germinated
6 coating_a germinated
7 coating_a germinated
8 coating_a germinated
9 coating_a germinated
10 coating_a germinated
# ℹ 110 more rows\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
observed_statistic <-
germ_data |>
specify(response = germination_success, explanatory = treatment) |>
hypothesize(null = "independence") |>
calculate(stat = "Chisq")
observed_statisticResponse: germination_success (factor)
Explanatory: treatment (factor)
Null Hypothesis: independence
# A tibble: 1 × 1
stat
<dbl>
1 2.43\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
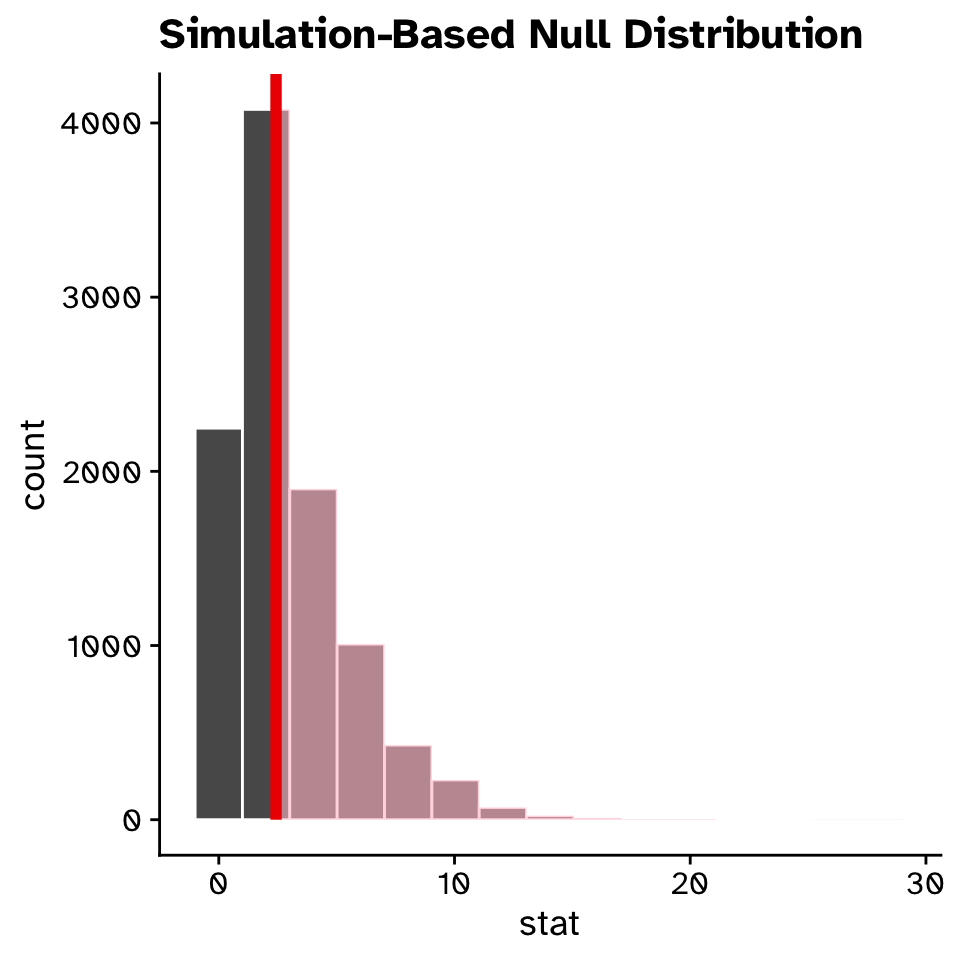
\(\chi^2\) Test of independence
Are two categorical variables associated with each other?
# A tibble: 1 × 1
p_value
<dbl>
1 0.906